#
Dr. M. Baron, Statistical Machine Learning class, STAT-427/627
# DIMENSION REDUCTION AND SHRINKAGE
Part II. Ridge Regression and LASSO
3. LASSO and RIDGE REGRESSION in package GLMNET
> library(glmnet)
# This package requires X-variables in a matrix
> X = model.matrix( medv ~ ., data=Boston )
> Y = medv
> ridgereg = glmnet(X,
Y, alpha=0, lambda = seq(0,10,0.01))
# alpha is a “mixing parameter”. It combines Lasso and Ridge
Regression. We only need the extreme values for now,
alpha=0 => ridge regression
alpha=1 => lasso
# So, which lambda is it best to choose? Run cross-validation…
> cv_ridge = cv.glmnet(X,medv,alpha=0,lambda=seq(0,10,0.01))
> names(cv_ridge)
[1]
"lambda" "cvm"
"cvsd" "cvup" "cvlo" "nzero" "name" "glmnet.fit"
[9]
"lambda.min" "lambda.1se"
# For the selected values of lambda, we get
“cvm” =
the mean cross-validation error
“cvsd” = its estimated standard error
“cvlo” =
cvm – cvsd (lower curve)
“cvup”= cvm + cvsd (upper curve)
# All these can be plotted…
> plot(cv_ridge)
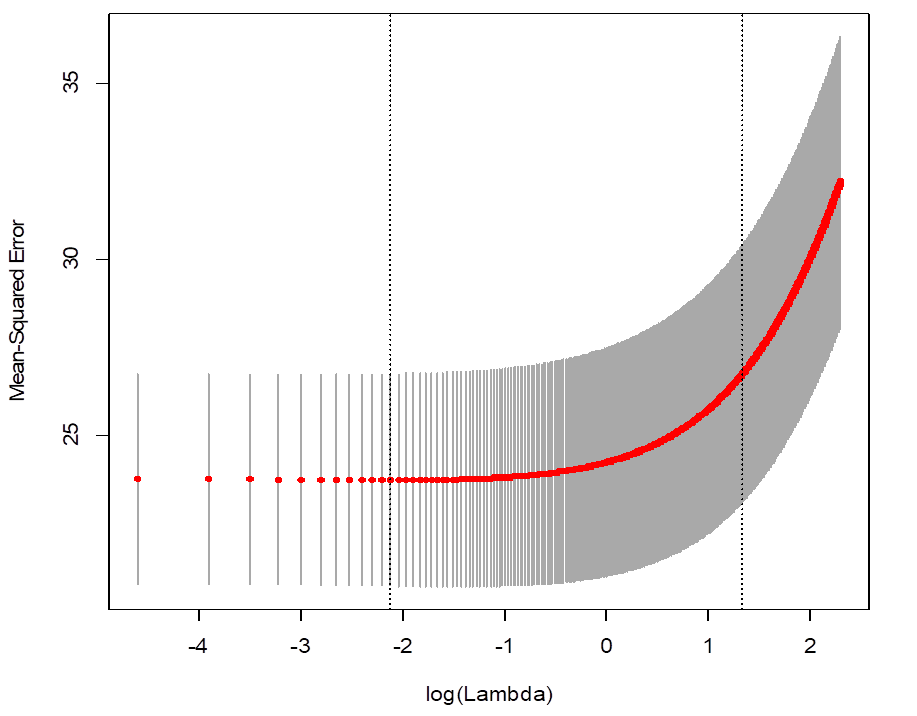
# Which lambda minimized the MSE?
> cv_ridge$lambda.min
[1] 0.12
> predict( ridgereg, cv_ridge$lambda.min,
type="coefficients" )
(Intercept)
20.863586526
crim -0.068099108
zn 0.020536577
indus -0.068164404
chas 2.777651593
nox -5.514335229
rm 3.645249869
age -0.007642606
dis -0.532994082
rad 0.034679405
tax -0.002916331
ptratio -0.676296397
black 0.007674913
lstat -0.351786530
# Similarly with LASSO, only choose alpha=1
> lasso = glmnet(X,
Y, alpha=1, lambda
= seq(0,10,0.01))
# Compare the slopes estimated by ridge regression and by lasso
> plot(ridgereg)
> plot(lasso)
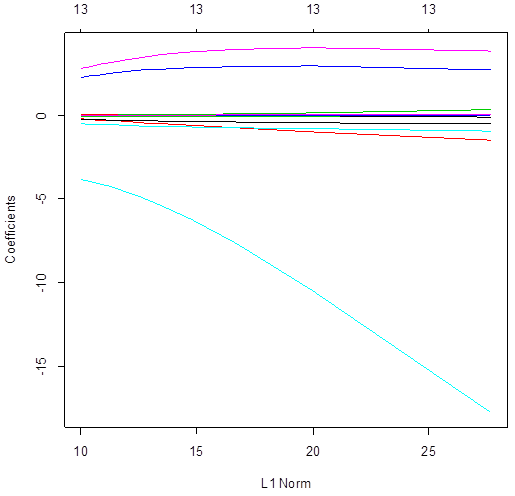
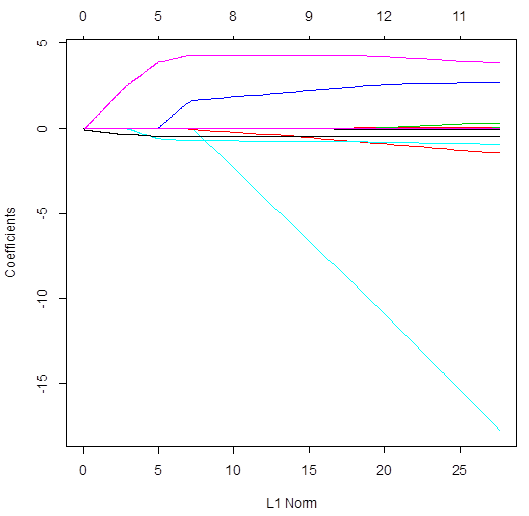
# Ridge regression uses all the variables – all slopes are not 0 for
all lambda. Conversely, lasso does variable selection
and sends some slopes to 0. The number of non-zero slopes is printed in the top.
> cv.lasso = cv.glmnet( X, medv, alpha=1, lambda=seq(0,10,0.01)
)
> plot(cv.lasso)
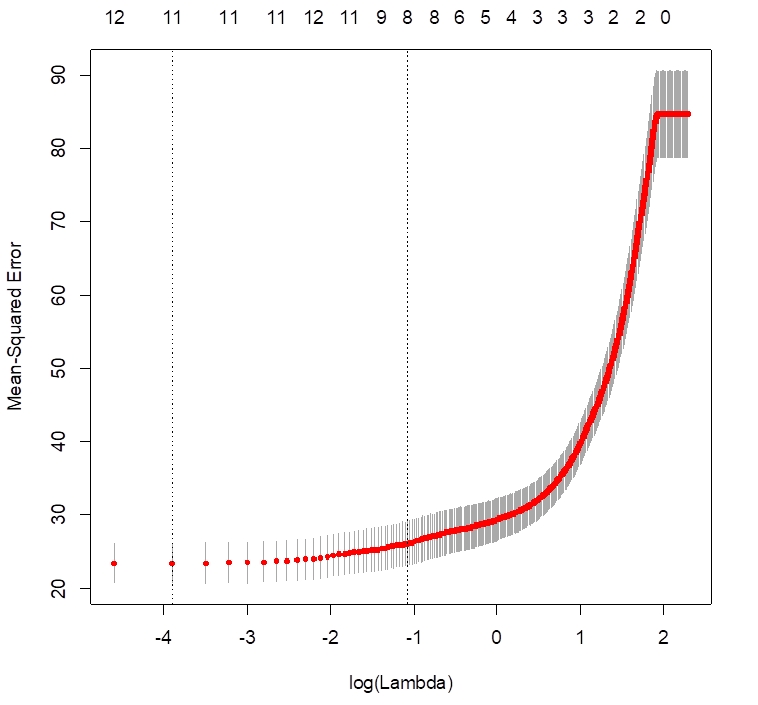
> cv.lasso$lambda.min
[1] 0.02
> predict( lasso,cv.
lasso$lambda.min, type="coefficients" )
(Intercept)
18.739101467
crim -0.024356546
zn .
indus .
chas 2.009577446
nox -4.667589527
rm 4.273554725
age .
dis -0.401567952
rad .
tax .
ptratio -0.803881292
black 0.006716721
lstat -0.518576315
# For LASSO, the best lambda to use is 0.02. Some coefficients are 0
– these variables are removed from the model.
# Prediction for new values of X and cross-validation
> n = length(medv)
> Z = sample(n,n/2)
> lasso = glmnet( X[Z,],
medv[Z], alpha=1, lambda=seq(0,10,0.01) )
> Yhat = predict( lasso,
cv.lasso$lambda.min, newx=X[-Z,] )
> mean((Yhat - medv[-Z])^2)
[1] 23.62894
# This is the test MSE, estimated by the validation set approach.